
Improved
Tumor Diagnosis
Next generation tumor boards

Predicting tumor chemosensitivity and relapse risk using Deep Learning
Our solution revolutionizes the prediction of response to treatment and the risk of relapse, bringing a considerable advance to current medical practices.
By combining the latest technological innovations with morphological analysis of tumors via Deep Learning algorithms, we offer a high-performance, accessible and easily integrable approach to more precise and personalized diagnosis. Join us in the transformation of diagnostic medicine and guarantee every patient the optimal treatment from day one.

Current challenges
Histological analysis remains the cornerstone of cancer treatment, providing precise identification of tumors through rigorous microscopic examination. This process is essential for adjusting treatments and ensuring optimal patient follow-up. However, traditional morphological classifications, such as SBR grading or AJCC stage for breast cancer (Valderrama et al., 2024), do not always reflect the actual response to treatment, thus limiting the chances of therapeutic success for some patients.
Classical approaches
Next-generation sequencing (NGS) and gene panels, applied to tumors or liquid biopsies, have transformed the management of cancer patients. These cutting-edge methods offer precise, personalized data to improve therapeutic outcomes (Smith et al., 2022; Johnson et al., 2023). However, they neglect an essential dimension: the histological analysis of tissues. Despite their innovation, these tools remain costly (several thousand euros per test), are limited by the small amount of tumor material available, and still present challenges in terms of accuracy and reliability (Norton et al., 2023).
Solution
Prediction of chemosensitivity, DFS (Disease Free Survival) and OS (Overall Survival) from AI histology analysis.
Accessibility
Can be performed on the basis of the initial diagnostic examination.
Economical
Just a few hundred euros.
Performance
Complete medical reporting to offer cutting-edge customized medicine from the day of diagnosis.
Our Technology
State-of-the-art Deep Learning
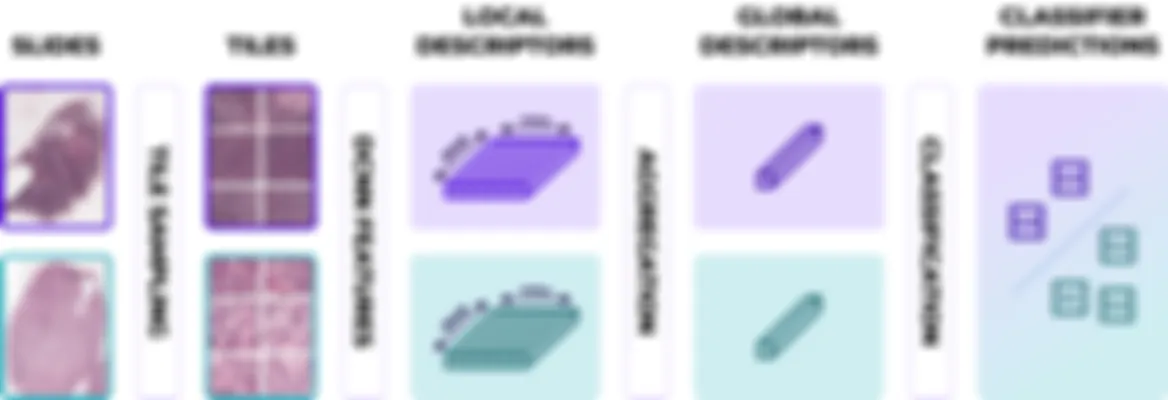
Chemo-prAIdict combines state-of-the-art proprietary algorithms based on weakly supervised and self supervised deep learning, combined with traditional machine learning and contextual molecular pathways, ensuring performances and fine-grained predictions for high precision diagnostics. (Valderrama et al. 2024)
Robustness
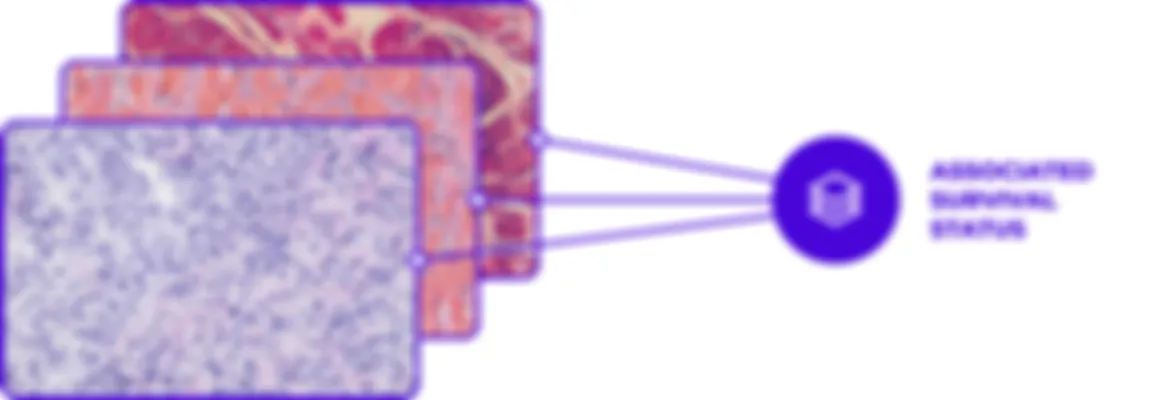
Our algorithms undergo rigorous testing on extensive external cohorts to ensure their reliability and accuracy. They are evaluated for biological consistency, which involves predicting patient outcomes using different samples to account for tumor heterogeneity.
Additionally, our algorithms are assessed for technical variability by being tested on different scanners, confirming their scanner-agnostic capabilities and ensuring robust technical correlation. This comprehensive validation process underscores our commitment to delivering precise and consistent diagnostic solutions.
Expert control
Chemo-prAIdict’s ergonomic design allows even first-time users to confidently manage the system, enabling quick and efficient analysis without compromising accuracy. Ideal for seamless integration into medical professionals’ diagnostic workflow.
Performance of our solution in predicting chemosensitivity(1)
OR
1
Standard classification
Not significant
OR
2.25-4.48
Genomic signature
(MammaPrint, OncotypeDx)(2)
OR
20.56
Chemo-prAIdict
(5-10 times more powerful than molecular signatures)
Breast-NEOprAIdict
The first software in our Chemo-prAIdict range covers all early breast cancers treated with neoadjuvant chemotherapy (HER2+, HER2-/ER+, TN). Compared with standard classification (molecular, AJCC, SBR), our biopsy analysis solution is extremely powerful:
| Methods | OR (95% CI) | AUC | P |
|---|---|---|---|
| HER2+ | 2.70 (1.08 - 6.76) | 0.67 | 0.0358 |
| HER2-/RH+ | 20.56 (1.14-371.74) | 0.87 | 0.00413 |
| TNBC | 3.02 (1.18-7.74) | 0.71 | 0.0206 |
We were able to test our solution on a cohort (compared with the standard classification):
- for HER2+ tumors: 3.63 more chances of detecting chemosensitive tumors.
- for TNBC: 2.03 more chances of detecting chemosensitive tumors.
- for luminal tumours (ER+/HER2-): 20.56 more chances of detecting the chemosensitive tumour.
These excellent results are the subject of further studies to further improve our performance and our level of clinical proof. We have also been able to initiate development programs in other pathologies, such as ovarian cancer and Hodgkin’s lymphoma.
(1) in early luminal breast cancer treated with neoadjuvant chemotherapy.
(2) Source: Freeman, J. Q. et al. Evaluation of multigene assays as predictors for response to neoadjuvant chemotherapy in early-stage breast cancer patients. npj Breast Cancer 9, 1–4 (2023)